The key to NGS Success
NGS multiplexing allows the simultaneous sequencing of multiple samples in a single run by using unique barcodes, significantly increasing throughput, and reducing costs while maintaining data quality and accuracy. NGS library quantification and NGS library normalization prior to pooling are imperative.
NGS library quantification refers to the process of accurately determining the concentration and quality of a DNA or RNA library prepared for sequencing. Quantification assays are essential at this stage to ensure libraries meet platform requirements.
NGS normalization is the process of adjusting the different concentrations of libraries to a uniform level before sequencing, ensuring equal representation of each library. These steps are crucial for ensuring optimal sequencing results, as they directly influence the efficiency and quality of the sequencing run. Myra supports library pooling NGS workflows through automation and precision in these critical tasks.
Importance of NGS Library Quantification and Normalization
Accurate Loading:
Proper NGS quantification and normalization ensure that the correct amount of library is loaded onto the sequencing platform. Overloading or underloading can lead to poor sequencing quality, reduced data yield, and increased costs.
Consistency:
Accurate quantification helps maintain consistency between different sequencing runs, enabling a reliable comparison of results.
Efficiency:
Proper quantification minimizes the need for reruns and optimizes the use of sequencing reagents and equipment, thus saving time and resources.
Fluourometric Quantifcation
Uses fluorescent dyes that bind specifically to double-stranded DNA (dsDNA). The fluorescence intensity is measured and correlated to DNA concentration. The Qubit Fluorometer is the most used instrument. The method is specific for dsDNA, reducing the impact of contaminants such as RNA or single-stranded DNA and can detect low concentrations of DNA. However, some fluorescent dyes can be inhibited by contaminants present in the sample
Capillary Electrophoresis (CE):
Separates DNA fragments based on their size and measures the fluorescence intensity of labelled fragments to determine concentration and size distribution. Examples such as the Agilent Bioanalyzer or Qsep Plus are typically used. The method provides information on both the concentration and size distribution of the library, which is also important to know. The method is not as accurate or sensitive as others, and the equipment and consumables can be expensive, not to mention time-consuming.
Quantitative PCR (qPCR):
Quantifies the amount of DNA by amplifying the specific adapter sequences of the library and measuring the accumulation of the amplified product in real-time. Examples like the NEB NGS Library Quantification Kit are used. Due to its high accuracy, sensitivity, and wide dynamic range, it is considered the Gold Standard of NGS Library quantification, and we would recommend using it in conjunction with CE.
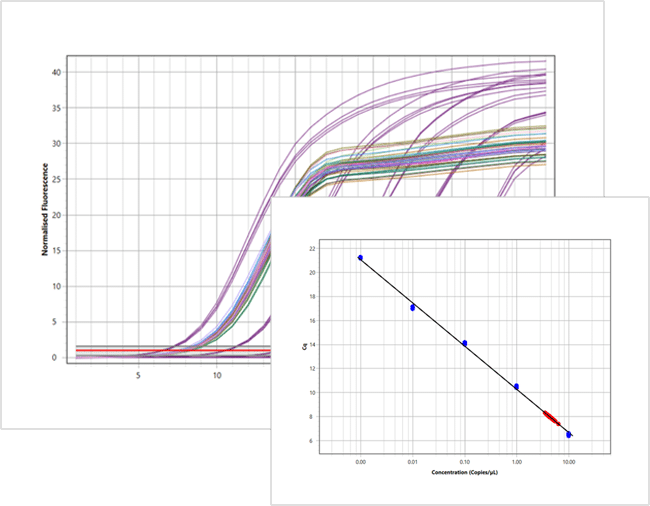
Myra Advantages for Normalization and Pooling
The Myra liquid handling system is highly regarded for its precision and reliability in setting up qPCR for NGS library quantification. Compared to other platforms, Myra produces superior results, making it a preferred choice for NGS library normalization and library pooling NGS tasks. One of the key advantages of using Myra is its level sensing feature, which can detect air pockets in wells – a common challenge that other platforms often struggle with. This capability ensures that each sample is pooled accurately, minimizing the risk of sample drop-outs.
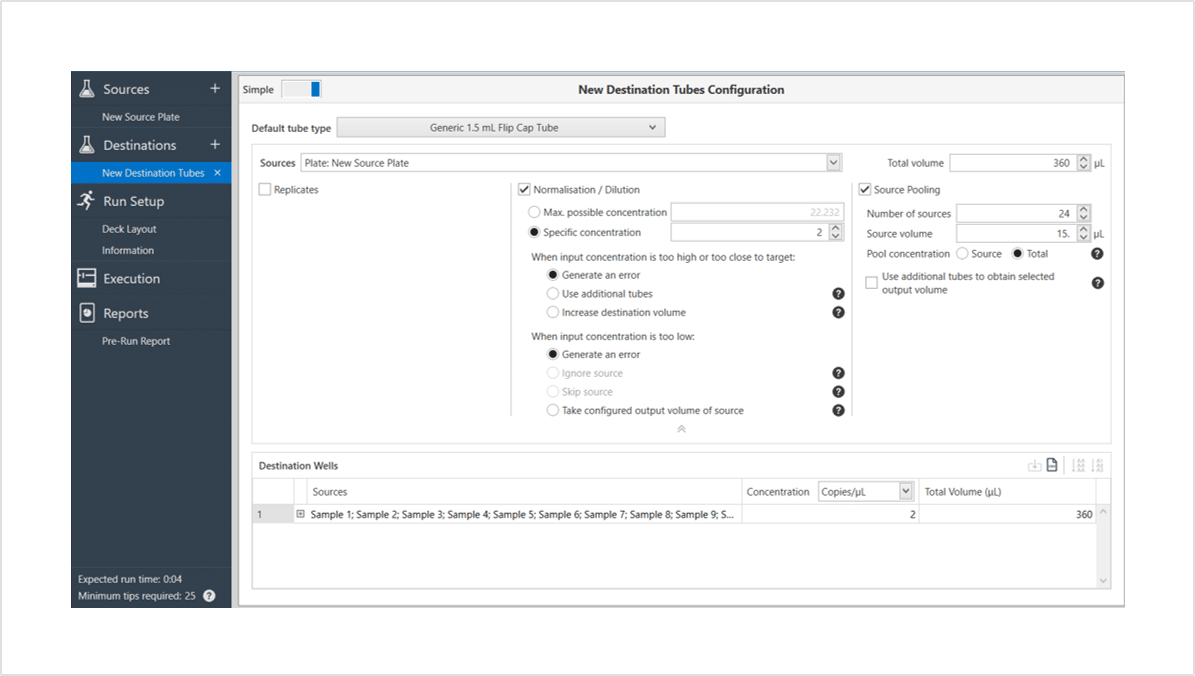
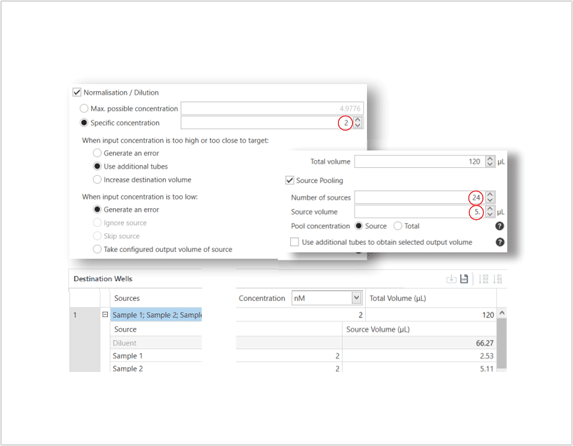
Setting up normalization and pooling on using the Workbench software is a simple as four easy steps:
- Import of enter your sample names and concentrations.
- Select the concentration to normalise samples to. The software will calculate a maximum value for you.
- Choose the number of samples per pool.
- Enter the volume of each sample or total for the pool.
The software will calculate the rest for you. It also has smart features to help determine how to handle samples that might be too high or too low to be normalised. Say goodbye to having to calculate in Excel.
Normalization and Pooling on Myra
Browse a comprehensive table of all available templates on Myra, complete with direct links to specific MyraScript pages. Access protocol documentation via a dedicated PDF link. Verified options include templates for BMS, Partner (kit company), and the Myra community (customers).
| Kit | Verified By |
|---|---|

| BMSCommunity |

| BMSCommunity |

| BMS |

| BMS |
Case Study: Normalization and Pooling by Ramaciotti Centre for Genomics
A Strong Preference For Myra
The Ramaciotti Centre for Genomics is a leading Australian research facility that provides advanced genomics services, including whole-genome sequencing, transcriptome analysis, and single-cell sequencing, utilizing state-of-the-art technologies to support diverse research needs in medicine, agriculture, and environmental science. Being one of the country’s leading sequencing service providers means the Ramaciotti Centre are tasked with providing high-quality sequencing reads for all their clients. Failure to achieve sufficient sequencing data can result in lost time, loss of precious sample and additional costs to re-run the sequencing. Here we provide an example of how the Ramaciotti Centre utilise Myra for all their normalization and pooling of whole genome library samples prior to running them on highly multiplexed sequencers such as the Illumina NovaSeq X. We show why Myra earns their trust and has become their go-to instrument.
Experimental Setup
Human genomic DNA samples were processed using the Illumina DNA library prep kit for use in whole genome sequencing. For each run, 24 samples were normalised and pooled using Myra to a starting concentration of 2 nM. The pooled samples were run on the Illumina NovaSeq X Plus platform. The read depth for each sample was analysed post-sequencing as part of the quality control for each run. A total of 4 runs were conducted for this study.
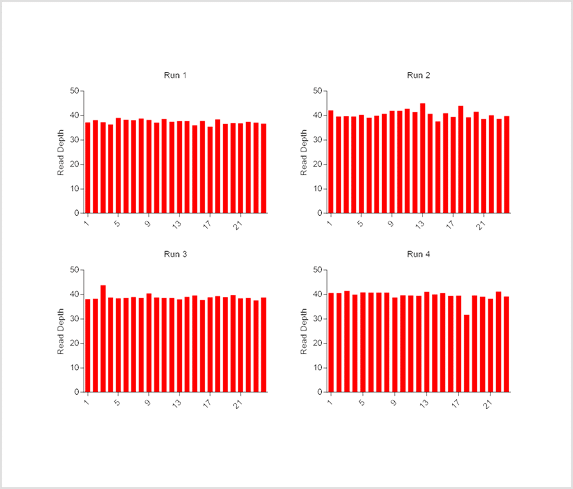
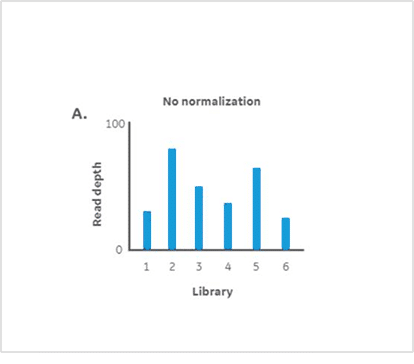
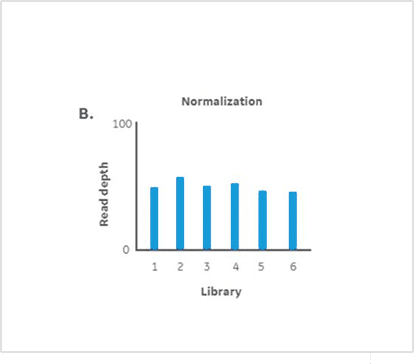
Results
Statistically consistent read depths were demonstrated in each of four separate sequencing runs following normalization and pooling using Myra (Figure 3). This indicates the reliable and reproducible performance of Myra in normalising and pooling samples for multiplexed NGS. The Ramaciotti Centre trusts Myra over other liquid handlers for consistently reliable results. Unlike other liquid handlers that require constant monitoring to ensure correct volume transfers, Myra uses a pressure-based liquid level sensing system to effectively normalise samples and trigger warnings if issues arise. This added assurance ensures accuracy and consistency in sample handling, maintaining high standards of data integrity. Taking into consideration that each highly multiplexed NovaSeq X run can cost well above $10,000 USD; any lost data can quickly become a financial burden for a sequencing service provider. Not to mention the lost time each client would experience along with their important samples. Myra’s pipetting accuracy and ease of use give the Ramaciotti group the peace of mind they need to continue their valuable work today and into the future.
“The Myra provides the perfect combination of speed and reliably that is lacking in many other platforms. You can trust that each experiment will run as planned and expected.” – Ramacoitti (NSW, Australia)
